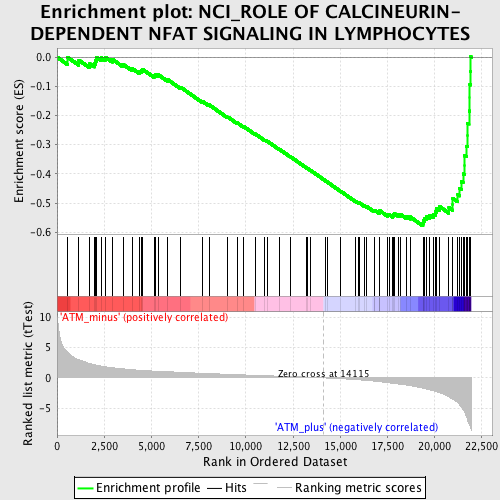
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | NCI_ROLE OF CALCINEURIN-DEPENDENT NFAT SIGNALING IN LYMPHOCYTES |
| Enrichment Score (ES) | -0.57850605 |
| Normalized Enrichment Score (NES) | -2.0694828 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0031622786 |
| FWER p-Value | 0.012 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CDK4 | 1422441_x_at | 544 | 4.192 | -0.0001 | No | ||
| 2 | XPO1 | 1418442_at 1418443_at 1448070_at | 1151 | 2.956 | -0.0103 | No | ||
| 3 | KPNB1 | 1416925_at 1434357_a_at 1448526_at 1451967_x_at | 1733 | 2.357 | -0.0229 | No | ||
| 4 | MAF | 1435828_at 1437473_at 1444073_at 1447849_s_at 1447945_at 1456060_at | 1970 | 2.170 | -0.0209 | No | ||
| 5 | PPARG | 1420715_a_at | 2038 | 2.118 | -0.0114 | No | ||
| 6 | IL2RA | 1420691_at 1420692_at | 2087 | 2.083 | -0.0013 | No | ||
| 7 | FOSL1 | 1417487_at 1417488_at | 2333 | 1.933 | -0.0010 | No | ||
| 8 | E2F1 | 1417878_at 1431875_a_at 1447840_x_at | 2542 | 1.816 | 0.0002 | No | ||
| 9 | SLC3A2 | 1425364_a_at | 2914 | 1.676 | -0.0069 | No | ||
| 10 | YWHAG | 1420816_at 1420817_at 1450012_x_at 1459693_x_at | 3489 | 1.477 | -0.0244 | No | ||
| 11 | CASP3 | 1426165_a_at 1430192_at 1449839_at | 3970 | 1.341 | -0.0384 | No | ||
| 12 | CALM3 | 1426710_at 1438825_at 1438826_x_at 1450864_at | 4358 | 1.257 | -0.0487 | No | ||
| 13 | EGR4 | 1449977_at | 4461 | 1.237 | -0.0460 | No | ||
| 14 | RNF128 | 1418318_at 1449036_at | 4517 | 1.228 | -0.0412 | No | ||
| 15 | CALM2 | 1422414_a_at 1423807_a_at | 5136 | 1.110 | -0.0630 | No | ||
| 16 | YWHAB | 1420878_a_at 1420879_a_at 1420880_a_at 1436783_x_at 1438708_x_at 1455815_a_at | 5228 | 1.094 | -0.0606 | No | ||
| 17 | IFNG | 1425947_at | 5363 | 1.070 | -0.0604 | No | ||
| 18 | RAN | 1443131_at 1446819_at 1447478_at 1460551_at | 5842 | 1.004 | -0.0764 | No | ||
| 19 | PRKCZ | 1418085_at 1449692_at 1454902_at | 6548 | 0.906 | -0.1033 | No | ||
| 20 | IL3 | 1450566_at | 7697 | 0.750 | -0.1513 | No | ||
| 21 | BAD | 1416582_a_at 1416583_at | 8047 | 0.703 | -0.1631 | No | ||
| 22 | MAPK14 | 1416703_at 1416704_at 1426104_at 1442364_at 1451927_a_at 1459617_at | 9038 | 0.581 | -0.2050 | No | ||
| 23 | TBX21 | 1449361_at | 9537 | 0.523 | -0.2247 | No | ||
| 24 | YWHAZ | 1416102_at 1416103_at 1436971_x_at 1436981_a_at 1439005_x_at 1443893_at 1448218_s_at 1448219_a_at | 9868 | 0.485 | -0.2369 | No | ||
| 25 | IL4 | 1449864_at | 10496 | 0.418 | -0.2631 | No | ||
| 26 | AKAP5 | 1447301_at | 11001 | 0.361 | -0.2840 | No | ||
| 27 | CSF2 | 1427429_at | 11147 | 0.346 | -0.2886 | No | ||
| 28 | TNF | 1419607_at | 11771 | 0.274 | -0.3155 | No | ||
| 29 | YWHAQ | 1420828_s_at 1420829_a_at 1420830_x_at 1432842_s_at 1437608_x_at 1454378_at 1460590_s_at 1460621_x_at | 12338 | 0.217 | -0.3401 | No | ||
| 30 | MAPK8 | 1420931_at 1420932_at 1437045_at 1440856_at 1457936_at | 13226 | 0.116 | -0.3800 | No | ||
| 31 | JUN | 1417409_at 1448694_at | 13265 | 0.110 | -0.3811 | No | ||
| 32 | PTGS2 | 1417262_at 1417263_at | 13435 | 0.089 | -0.3883 | No | ||
| 33 | SFN | 1448612_at | 14234 | -0.019 | -0.4247 | No | ||
| 34 | PTPRK | 1423277_at 1423278_at 1431680_a_at 1441276_at 1441277_s_at 1441644_at 1443491_at 1443699_at 1444763_at | 14337 | -0.034 | -0.4292 | No | ||
| 35 | PRKCE | 1437860_at 1437861_s_at 1442609_at 1444649_at 1449956_at 1452878_at | 15005 | -0.138 | -0.4589 | No | ||
| 36 | PIM1 | 1423006_at 1435458_at | 15787 | -0.279 | -0.4930 | No | ||
| 37 | PRKCA | 1427562_a_at 1443069_at 1445028_at 1445395_at 1446598_at 1450945_at | 15983 | -0.319 | -0.5000 | No | ||
| 38 | FKBP1A | 1416036_at 1438795_x_at 1438958_x_at 1448184_at 1456196_x_at | 16008 | -0.325 | -0.4992 | No | ||
| 39 | IRF4 | 1421173_at 1421174_at | 16277 | -0.392 | -0.5091 | No | ||
| 40 | CD40LG | 1422283_at | 16403 | -0.421 | -0.5123 | No | ||
| 41 | NFATC1 | 1417621_at 1425761_a_at 1428479_at 1447084_at 1447085_s_at | 16786 | -0.526 | -0.5267 | No | ||
| 42 | PRKACA | 1447720_x_at 1450519_a_at | 16824 | -0.536 | -0.5252 | No | ||
| 43 | FOXP3 | 1420765_a_at 1457082_at | 17059 | -0.612 | -0.5323 | No | ||
| 44 | PTPN1 | 1417068_a_at 1438670_at | 17061 | -0.612 | -0.5287 | No | ||
| 45 | FASLG | 1418803_a_at 1449235_at | 17094 | -0.623 | -0.5265 | No | ||
| 46 | CALM1 | 1417365_a_at 1417366_s_at 1433592_at 1454611_a_at 1455571_x_at | 17505 | -0.779 | -0.5406 | No | ||
| 47 | POU2F1 | 1419716_a_at 1427695_a_at 1427835_at | 17578 | -0.808 | -0.5391 | No | ||
| 48 | MAPK9 | 1421876_at 1421877_at 1421878_at | 17782 | -0.895 | -0.5431 | No | ||
| 49 | CSNK1A1 | 1424827_a_at 1428537_at 1430529_at 1451497_at 1456216_at | 17814 | -0.908 | -0.5392 | No | ||
| 50 | MEF2D | 1421388_at 1434487_at 1436642_x_at 1437300_at 1458901_at | 17877 | -0.936 | -0.5365 | No | ||
| 51 | CAMK4 | 1421941_at 1426167_a_at 1439843_at 1442822_at 1452572_at | 18098 | -1.010 | -0.5406 | No | ||
| 52 | FKBP8 | 1416113_at | 18186 | -1.035 | -0.5384 | No | ||
| 53 | NUP214 | 1434351_at 1456503_at 1457815_at | 18505 | -1.172 | -0.5460 | No | ||
| 54 | MAP3K8 | 1419208_at | 18698 | -1.260 | -0.5473 | No | ||
| 55 | MAPK3 | 1427060_at | 19380 | -1.691 | -0.5685 | Yes | ||
| 56 | IL5 | 1425546_a_at 1442049_at 1450550_at | 19394 | -1.700 | -0.5590 | Yes | ||
| 57 | PRKCH | 1422079_at 1434248_at | 19474 | -1.756 | -0.5522 | Yes | ||
| 58 | NFATC2 | 1425901_at 1425990_a_at 1426031_a_at 1426032_at 1439205_at 1440426_at | 19581 | -1.851 | -0.5461 | Yes | ||
| 59 | YWHAE | 1426384_a_at 1426385_x_at 1435702_s_at 1438839_a_at 1440841_at 1457173_at | 19732 | -1.981 | -0.5413 | Yes | ||
| 60 | CBLB | 1437304_at 1455082_at 1458469_at | 19947 | -2.123 | -0.5385 | Yes | ||
| 61 | YWHAH | 1416004_at | 20026 | -2.200 | -0.5290 | Yes | ||
| 62 | ITCH | 1415769_at 1431316_at 1459332_at | 20078 | -2.248 | -0.5181 | Yes | ||
| 63 | CTLA4 | 1419334_at | 20241 | -2.409 | -0.5112 | Yes | ||
| 64 | GATA3 | 1448886_at | 20751 | -3.097 | -0.5162 | Yes | ||
| 65 | FOS | 1423100_at | 20955 | -3.496 | -0.5048 | Yes | ||
| 66 | EP300 | 1434765_at | 20959 | -3.504 | -0.4842 | Yes | ||
| 67 | CREBBP | 1434633_at 1435224_at 1436983_at 1444856_at 1457641_at 1459804_at | 21224 | -4.102 | -0.4720 | Yes | ||
| 68 | BCL2 | 1422938_at 1427818_at 1437122_at 1440770_at 1443837_x_at 1457687_at | 21309 | -4.363 | -0.4500 | Yes | ||
| 69 | PRKCQ | 1426044_a_at | 21404 | -4.723 | -0.4263 | Yes | ||
| 70 | JUNB | 1415899_at | 21503 | -5.184 | -0.4001 | Yes | ||
| 71 | NR4A1 | 1416505_at | 21561 | -5.542 | -0.3699 | Yes | ||
| 72 | MAP3K1 | 1424850_at | 21563 | -5.558 | -0.3370 | Yes | ||
| 73 | NFATC3 | 1419976_s_at 1452497_a_at | 21671 | -6.290 | -0.3047 | Yes | ||
| 74 | PRKCD | 1422847_a_at 1442256_at | 21741 | -6.917 | -0.2669 | Yes | ||
| 75 | DGKA | 1418578_at | 21744 | -6.953 | -0.2258 | Yes | ||
| 76 | CSNK2A1 | 1419034_at 1419035_s_at 1419036_at 1419037_at 1419038_a_at 1453427_at | 21816 | -7.560 | -0.1843 | Yes | ||
| 77 | EGR1 | 1417065_at | 21860 | -7.832 | -0.1399 | Yes | ||
| 78 | GSK3B | 1434439_at 1437001_at 1439931_at 1439949_at 1451020_at 1454958_at | 21861 | -7.833 | -0.0936 | Yes | ||
| 79 | EGR2 | 1427682_a_at 1427683_at | 21904 | -8.113 | -0.0475 | Yes | ||
| 80 | EGR3 | 1421486_at 1436329_at | 21913 | -8.258 | 0.0011 | Yes |

